TypeError: '(slice(None, None, None), 0)' is an invalid key
Last updated: Apr 13, 2024
Reading time·3 min
# TypeError: '(slice(None, None, None), 0)' is an invalid key
The Python "TypeError: '(slice(None, None, None), 0)' is an invalid key"
occurs when you try to directly slice a DataFrame using extended NumPy array
slicing.
To solve the error, use the DataFrame.iloc attribute or convert the input
from a DataFrame to a NumPy array using .to_numpy().
Here is an example of how the error occurs.
import numpy as np import pandas as pd from sklearn.impute import SimpleImputer imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean') imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) data = [ [np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9], ] X_train = pd.DataFrame(data=data) # ⛔ TypeError: '(slice(None, None, None), 0)' is an invalid key imputer = imp_mean.fit(X_train[:, 0:2]) X_train[:, 0:2] = imputer.transform(X_train[:, 0:2]) print(X_train)
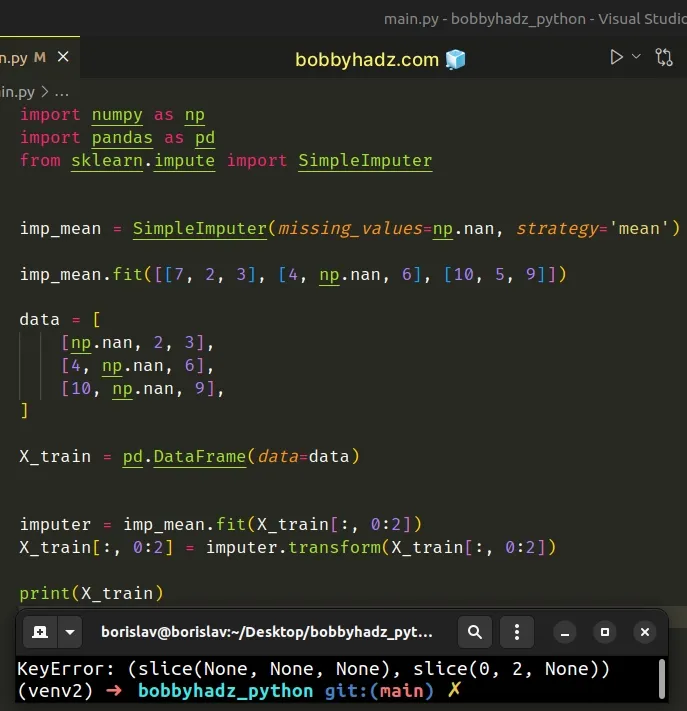
The error is caused because we are trying to directly slice a DataFrame using
extended NumPy array indexing.
# Using the DataFrame.iloc indexer to solve the error
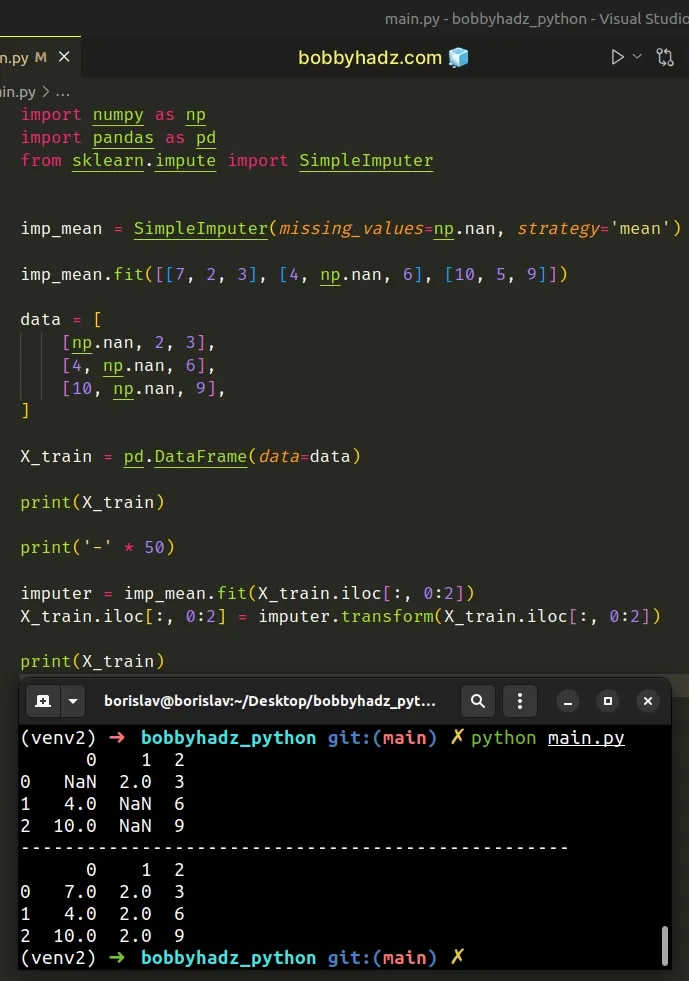
One way to solve the error is to use the DataFrame.iloc position-based indexer.
import numpy as np import pandas as pd from sklearn.impute import SimpleImputer imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean') imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) data = [ [np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9], ] X_train = pd.DataFrame(data=data) print(X_train) print('-' * 50) # ✅ Using `DataFrame.iloc` instead imputer = imp_mean.fit(X_train.iloc[:, 0:2]) X_train.iloc[:, 0:2] = imputer.transform(X_train.iloc[:, 0:2]) print(X_train)
Running the code sample produces the following output.
0 1 2 0 NaN 2.0 3 1 4.0 NaN 6 2 10.0 NaN 9 -------------------------------------------------- 0 1 2 0 7.0 2.0 3 1 4.0 2.0 6 2 10.0 2.0 9
The DataFrame.iloc attribute enables us to slice a DataFrame as shown in the
code sample.
X_train.iloc[:, 0:2] = imputer.transform(X_train.iloc[:, 0:2])
When slicing a DataFrame in your code, always make sure to use the .iloc
attribute.
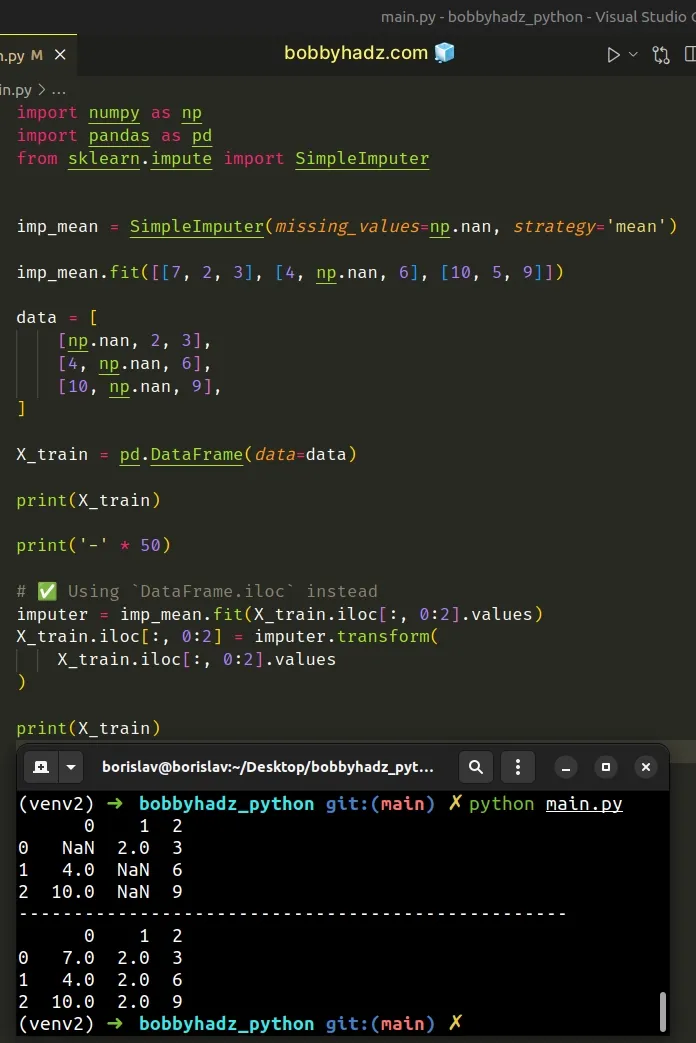
If the error persists, try to use DataFrame.iloc.[].values.
import numpy as np import pandas as pd from sklearn.impute import SimpleImputer imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean') imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) data = [ [np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9], ] X_train = pd.DataFrame(data=data) print(X_train) print('-' * 50) # ✅ Using `DataFrame.iloc[].values` instead imputer = imp_mean.fit(X_train.iloc[:, 0:2].values) X_train.iloc[:, 0:2] = imputer.transform(X_train.iloc[:, 0:2].values) print(X_train)
# Converting the DataFrame to a NumPy array to solve the error
Alternatively, you can solve the error by using the
DataFrame.to_numpy()
method to convert the input DataFrame to a NumPy array.
import numpy as np import pandas as pd from sklearn.impute import SimpleImputer imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean') imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) data = [ [np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9], ] X_train = pd.DataFrame(data=data) X_train = X_train.to_numpy() print(X_train) print('-' * 50) imputer = imp_mean.fit(X_train[:, 0:2]) X_train[:, 0:2] = imputer.transform(X_train[:, 0:2]) print(X_train)
Running the code sample produces the following output.
[[nan 2. 3.] [ 4. nan 6.] [10. nan 9.]] -------------------------------------------------- [[ 7. 2. 3.] [ 4. 2. 6.] [10. 2. 9.]]
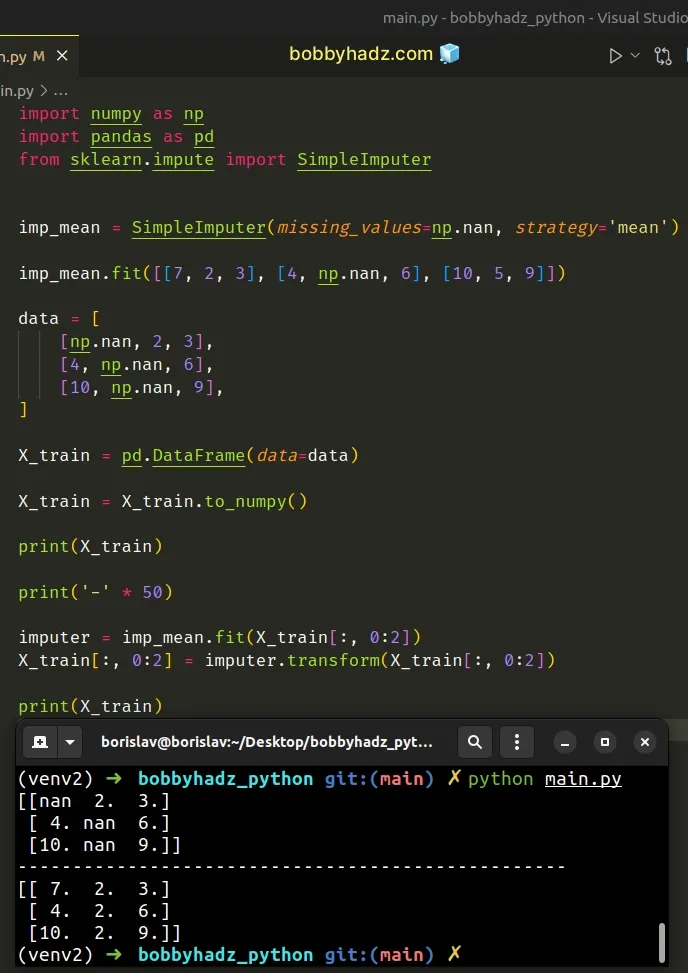
The DataFrame.to_numpy() method converts the DataFrame to a NumPy array.
X_train = X_train.to_numpy() # [[nan 2. 3.] # [ 4. nan 6.] # [10. nan 9.]] print(X_train)
# Additional Resources
You can learn more about the related topics by checking out the following tutorials:

